Automatic Differentiation
CSE 891: Deep Learning
Vishnu Boddeti
Implementing Backpropagation
- Implementing backpropagation by hand is like doing assembly programming.
- You don't have to do it most of the times.
- You need it sometimes, when desired functionality is absent.
- You do need it sometimes, if you want to eke out all the efficiencies out of the hardware.
- Can we automate the process of computing gradients of arbitrary functions?
- Autodiff: An automatic differentiation library.
Terminology
- Automatic Differentiation (AutoDiff): A general purpose solution for taking a program that computes a scalar value and automatically constructing a procedure for the computing the derivative of that value.
- Forward Mode Autodiff
- Reverse Mode Autodiff
- Backpropagation: A special case of autodiff applied to neural networks. The words "backpropagation" and "autodiff" are used interchangeably in machine learning.
- AutoGrad: It is the name of a particular autodiff package.
Finite Differences
- We often use finite differences to check our gradient calculations.
- One-sided version: $\begin{equation}\frac{\partial}{\partial x_i}f(x_1,\dots,x_N)\approx\frac{f(x_1,\dots,x_i+h,\dots,x_N)-f(x_1,\dots,x_i,\dots,x_N)}{h}\end{equation}$
- Two-sided version: $\begin{equation}\frac{\partial}{\partial x_i}f(x_1,\dots,x_N)\approx\frac{f(x_1,\dots,x_i+h,\dots,x_N)-f(x_1,\dots,x_i-h,\dots,x_N)}{2h}\end{equation}$

What Autodiff is not...
- Finite Difference Approximation
- Computationally expensive, needs many forward passes.
- Can induce large numerical errors.
- Normally, we only use it for testing.
- Symbolic Differentiation
- Can result in complex and redundant expressions.
- There may not be a convenient formula for the derivative.
What Autodiff is...
- An efficient (linear in the cost of computing the scalar value) and numerically stable way of computing gradients.
- A procedure for computing the derivatives, as opposed to obtaining a formula.
Recap: Derivative Example
- Forward Pass: $$ \begin{eqnarray} z &=& wx + b \nonumber \\ y &=& \sigma(z) \nonumber \\ \mathcal{L} &=& \frac{1}{2}(y-t)^2 \nonumber \end{eqnarray} $$
- Computing Derivatives using Chain Rule: $$ \begin{eqnarray} \mathcal{L}' &=& 1 \nonumber \\ y' &=& y - t \nonumber \\ z' &=& y'\sigma'(z) \nonumber \\ w' &=& z'x \nonumber \\ b' &=& z' \nonumber \end{eqnarray} $$
Autodiff Example Continued...
- Autodiff systems convert the program into a sequence of known primitive operations with known routines for computing derivatives.
- Under this representation, backpropagation reduces to a series of operations.
- Forward Pass: $$ \begin{eqnarray} z &=& wx + b \nonumber \\ y &=& \frac{1}{1+exp(-z)} \nonumber \\ \mathcal{L} &=& \frac{1}{2}(y-t)^2 \nonumber \end{eqnarray} $$
- Sequence of primitive operations:
$$
\begin{eqnarray}
t_1 &=& wx \nonumber \\
z &=& t_1 + b \nonumber \\
t_3 &=& -z \nonumber \\
t_4 &=& \exp(t_3) \nonumber \\
t_5 &=& 1 + t_4 \nonumber
\end{eqnarray}
$$
$$
\begin{eqnarray}
y &=& \frac{1}{t_5} \nonumber \\
t_6 &=& y - t \nonumber \\
t_7 &=& t_6^2 \nonumber \\
\mathcal{L} &=& \frac{t_7}{2} \nonumber
\end{eqnarray}
$$
Autodiff Example
import autograd.numpy as np
from autograd import grad
def sigmoid(x):
return 0.5*(np.tanh(x) + 1)
def logistic_predictions(weights, inputs):
# Outputs probability of a label being true according to logistic model.
return sigmoid(np.dot(inputs, weights))
def training_loss(weights):
# Training loss is the negative log-likelihood of the training labels.
preds = logistic_predictions(weights, inputs)
label_probabilities = preds * targets + (1 - preds) * (1 - targets)
return -np.sum(np.log(label_probabilities))
# Build a toy dataset.
inputs = np.array([[0.52, 1.12, 0.77],
[0.88, -1.08, 0.15],
[0.52, 0.06, -1.30],
[0.74, -2.49, 1.39]])
targets = np.array([True, True, False, True])
# Build a function that returns gradients of training loss using autograd.
training_gradient_fun = grad(training_loss)
# Check the gradients numerically, just to be safe.
weights = np.array([0.0, 0.0, 0.0])
# Optimize weights using gradient descent.
print("Initial loss:", training_loss(weights))
for i in range(100):
weights -= training_gradient_fun(weights) * 0.01
print("Trained loss:", training_loss(weights))
Autograd
- Autograd is a package that implements the ideas behind automatic differentiation.
- The original Autograd package can be found at:
- Autodidact: a simple and illustrative example of Autograd.
- JAX: successor of Autograd
Computational Graph
- Autodiff packages typically first explicitly construct the computational graph.
- Frameworks like Tensorflow provide custom language (API) for building computation graphs directly.
- Frameworks like PyTorch and Autograd instead build the computational graph by tracing all the operations during forward pass.
- The Node class represents a node of the computation graph. The attributes of the class are:
- value: actual value computed on a particular set of inputs
- fun: the primitive operation defining the node
- args: the arguments the op was called with
- parents: the parents of the current node
Computational Graph Continued...
- Autograd wraps basic Numpy ops while enabling it to build a computation graph.

Computational Graph Example
def logistic(z):
return 1./(1. + np.exp(-z))
# that is equivalent to:
def logistic2(z):
return np.reciprocal(np.add(1, np.exp(np.negative(z))))
z = 1.5
y = logistic(z)

Vector-Jacobian Products
- Reminder: Backpropagation equations can be expressed in terms of sums and indices.
- Goal: Implement primitive operations and backpropagation in vectorized form.
- The Jacobian is the matrix of partial derivatives: $$\begin{equation}\mathbf{J} = \frac{\partial \mathbf{y}}{\partial \mathbf{x}} = \begin{bmatrix} \frac{\partial y_1}{\partial x_1} & \dots & \frac{\partial y_1}{\partial x_n} \\\ \vdots & \ddots & \vdots \\\ \frac{\partial y_n}{\partial x_1} & \dots & \frac{\partial y_n}{\partial x_n} \end{bmatrix}\end{equation}$$
- Backpropagation equation at a given node can be written as a vector-Jacobian product (VJP): $$\begin{equation}x_j' = \sum_{i} y'_i \frac{\partial y_i}{\partial x_j}\end{equation}$$
- We can re-write this as a column vector by taking: $\mathbf{x}' = \mathbf{J}^T\mathbf{y}'$
VJP Examples
- Examples:
- Matrix Vector Product $$\begin{equation}\mathbf{z} = \mathbf{W}\mathbf{x} \mbox{ and } \mathbf{J} = \mathbf{W} \mbox{ and } \mathbf{x}' = \mathbf{W}^T\mathbf{z}'\end{equation}$$
- Elementwise Operations $$\begin{equation}\mathbf{y} = \exp(\mathbf{z}) \mbox{ and } \mathbf{J} = \begin{bmatrix} \exp(z_1) & & 0 \\\ & \ddots & \\\ 0 & & \exp(z_n) \end{bmatrix} \mbox{ and } \mathbf{z}' = \exp(\mathbf{z})\circ\mathbf{y}'\end{equation}$$
- In practice, we never explicitly construct the Jacobian, we instead directly compute the vector-Jacobian product for simplicity and efficiency.
VJP Examples Continued...
- For each primitive operation, we must specify VJPs for each of its arguments. Consider $y = \exp(x)$
- This is a function which takes in the output gradient (i.e. $y'$), the answer ($y$), and the arguments ($x$), and returns the input gradient ($x'$).
- defvjp (defined in core.py) is a convenience routine for registering VJPs. It just adds them to a dict.
- Here are some examples from numpy/numpy_vjps.py:
defvjp(negative, lambda g, ans, x: -g)
defvjp(exp, lambda g, ans, x: ans * g)
defvjp(log, lambda g, ans, x: g / x)
defvjp(add, lambda g, ans, x, y: g,
lambda g, ans, x, y: g)
defvjp(multiply, lambda g, ans, x, y: y * g,
lambda g, ans, x, y: x * g))
defvjp(subtract, lambda g, ans, x, y: g,
lambda g, ans, x, y: -g)
Backpropagation
- Backpropagation computations can be seen as message passing on the computational graph.
- This procedure can be directly implemented using an ordered list and modular functions.
Backpropagation as Message Passing

- Consider, standard backpropagation for computing $\mathbf{z}'$: $$\begin{equation}\mathbf{z}' = \frac{\partial \mathbf{r}}{\partial \mathbf{z}}\mathbf{r}' + \frac{\partial \mathbf{s}}{\partial \mathbf{z}}\mathbf{s}' + \frac{\partial \mathbf{t}}{\partial \mathbf{z}}\mathbf{t}'\end{equation}$$
- Not modular: $\mathbf{z}$ needs to know how it is used in the network for computing partial derivatives of $\mathbf{r}$, $\mathbf{s}$, and $\mathbf{t}$.
Backprop as Message Passing Continued...
- Node receives messages from children, aggregates them to get its own error signal, then passes messages to parents.
- Each message is a VJP

- Modularity: each node needs to know how to compute its outgoing messages, i.e. the VJPs corresponding to each of its parents.
- Implementation of $\mathbf{z}$ is agnostic to knowing where $\mathbf{z}'$ came from.
Backward Pass
- The backwards pass is defined in core.py.
- The argument $g$ is the error signal for the end node. This is always $\mathcal{L}' = 1$.
def backward_pass(g, end_node):
outgrads = {end_node : (g, False)}
for node in toposort(end_node):
outgrad = outgrads.pop(node)
ingrads = node.vjp(outgrad[0])
for parent, ingrad in zip(node.parents, ingrads):
outgrads[parent] = add_outgrads(outgrads.get(parent), ingrad)
return outgrad[0]
def add_outgrads(prev_g, g):
if prev_g is None:
return g
return prev_g + g
Backward Pass
- grad is just a wrapper around make_vjp which builds the computation graph and feeds it to backward_pass.
- grad itself is viewed as a VJP, if we treat $\bar{\mathcal{L}}$ as the $1 \times 1$ matrix with entry 1.
def make_vjp(fun, x):
start_node = VJPNode.new_root()
end_value, end_node = trace(start_node, fun, x)
if end_node is None:
def vjp(g): return vspace(x).zeros()
else:
def vjp(g): return backward_pass(g, end_node)
return vjp, end_value
def grad(fun, argnum=0):
def gradfun(*args, **kwargs):
unary_fun = lambda x: fun(*subval(args, argnum, x), **kwargs)
vjp, ans = make_vjp(unary_fun, args[argnum])
return vjp(np.ones_like(ans))
return gradfun
Automatic Differentiation Modes
- Consider: $$\mathcal{L} = F(G(H(x)))$$
- Forward-Mode Differentiation: $$ \begin{eqnarray} \frac{\partial \mathcal{L}}{\partial x} =\left(\frac{\partial \mathcal{L}}{\partial F}\left(\frac{\partial F}{\partial G}\left(\frac{\partial G}{\partial H}\frac{\partial H}{\partial x}\right)\right)\right) \nonumber \end{eqnarray} $$
- Reverse-Mode Differentiation: $$ \begin{eqnarray} \frac{\partial \mathcal{L}}{\partial x} = \left(\left(\left(\frac{\partial \mathcal{L}}{\partial F}\frac{\partial F}{\partial G}\right)\frac{\partial G}{\partial H}\right)\frac{\partial H}{\partial x}\right) \nonumber \end{eqnarray} $$
Reverse-Mode AD

- Matrix multiplication is associative: we can compute products in any order.
- Computing products right-to-left avoids matrix-matrix products; only needs matrix-vector $$\begin{equation}\frac{\partial \mathcal{L}}{\partial x_0} = \left(\frac{\partial x_1}{\partial x_0}\right)\left(\frac{\partial x_2}{\partial x_1}\right)\left(\frac{\partial x_3}{\partial x_2}\right)\left(\frac{\partial \mathcal{L}}{\partial x_3}\right)\end{equation}$$
Reverse Mode Automatic Differentiation
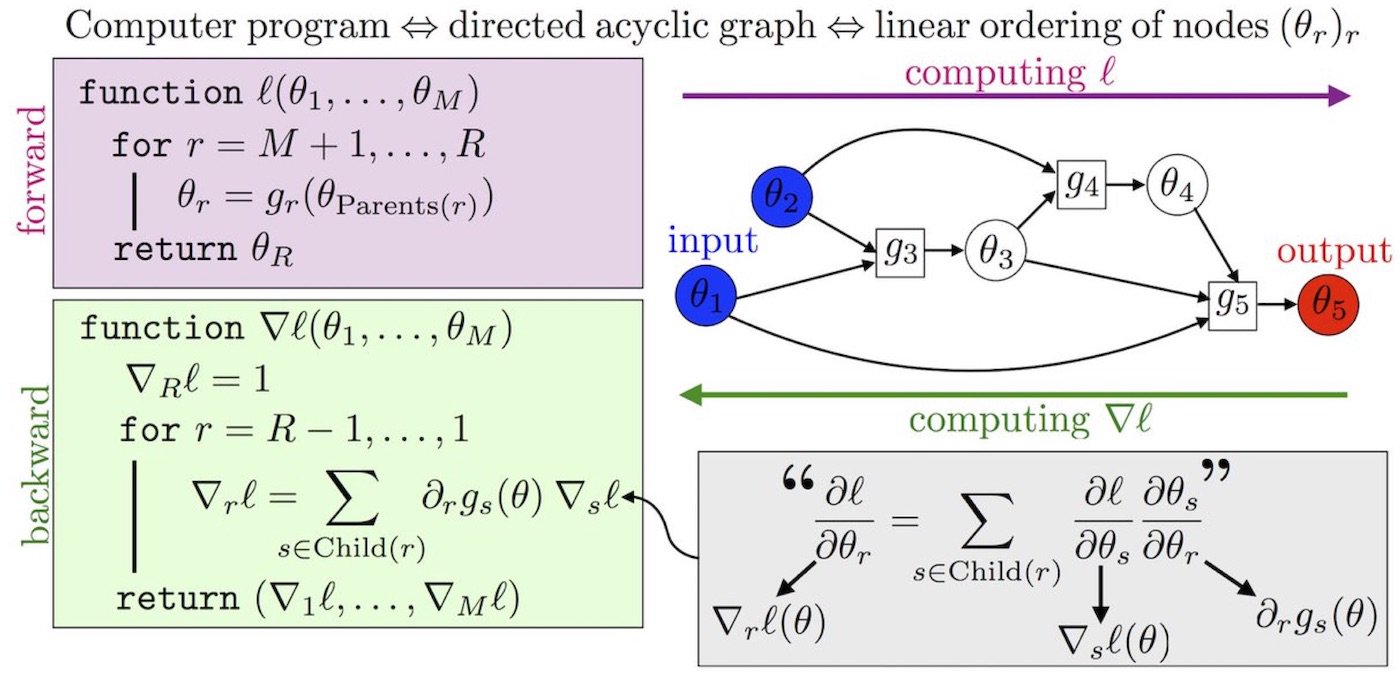
Forward-Mode AD

- Computing products left-to-right avoids matrix-matrix products; only needs matrix-vector
- Not implemented in Pytorch/Tensorflow $$\begin{equation}\frac{\partial \mathcal{L}}{\partial x_0} = \left(\frac{\partial x_1}{\partial x_0}\right)\left(\frac{\partial x_2}{\partial x_1}\right)\left(\frac{\partial x_3}{\partial x_2}\right)\left(\frac{\partial \mathcal{L}}{\partial x_3}\right)\end{equation}$$
Higher-Order Derivatives

- Hessian Matrix: second derivatives
- Hessian-vector multiply
$$\begin{equation}\frac{\partial^2 \mathcal{L}}{\partial \mathbf{x}^2_0}\mathbf{v}=\frac{\partial}{\partial \mathbf{x}_0}\left[\frac{\partial \mathcal{L}}{\partial \mathbf{x}_0}\cdot \mathbf{v}\right]\end{equation}$$
Higher-Order Derivatives Example

- Example: Regularization to penalize the norm of the gradient $$\begin{equation}R(\mathbf{W})=\|\frac{\partial \mathcal{L}}{\partial \mathbf{W}}\|_2^2=\left(\frac{\partial \mathcal{L}}{\partial \mathbf{W}}\right)\left(\frac{\partial \mathcal{L}}{\partial \mathbf{W}}\right) \frac{\partial }{\partial \mathbf{x}_0}\left[R(\mathbf{W})\right]=2\left(\frac{\partial^2\mathcal{L}}{\partial\mathbf{x}^2_0}\right)\left(\frac{\partial\mathcal{L}}{\partial\mathbf{x}_0}\right)\end{equation}$$
- Gulrajani et al, "Improved Training of Wasserstein GANs", NeurIPS 2017
Summary
- We saw three main parts to the code:
- tracing the forward pass to build the computation graph
- vector-Jacobian products for the primitive operations
- backward pass
- You are encouraged to read the full code ($< 200$ lines!) at: Autograd
Backpropagation Through Fluid Simulation
Autograd ExamplesHyperparameter Optimization Through Gradients

Hyperparameter Optimization Through Gradients

- Maclaurin, Dougal, David Duvenaud, and Ryan Adams. "Gradient-based hyperparameter optimization through reversible learning." ICML 2015
Hyperparameter Optimization Through Gradients Continued...

Learning to learning by gradient descent by gradient descent

Q & A