Graph Neural Networks - II
CSE 891: Deep Learning
Vishnu Boddeti
Recap: Graph Structured Data

Standard CNN and RNN architectures don't work on this data.
- Slide Credit: Xavier Bresson
Recap: Graph Definition
- Graphs $\mathcal{G}$ are defined by:
- Vertices $V$
- Edges $E$
- Adjacency matrix $\mathbf{A}$
- Features:
- Node features: $\mathbf{h}_i, \mathbf{h}_j$ (user type)
- Edge features: $\mathbf{e}_{ij}$ (relation type)
- Graph features: $\mathbf{g}$ (network type)

- We want our main desiderata, permutation invariance and permutation equivariance, to still hold
Node Classification

- Figure Credit: Jure Leskovec
Link Prediction

- Figure Credit: Jure Leskovec
Link Prediction: Social Networks

- Figure Credit: Michael Bronstein
Link Prediction: Social Networks
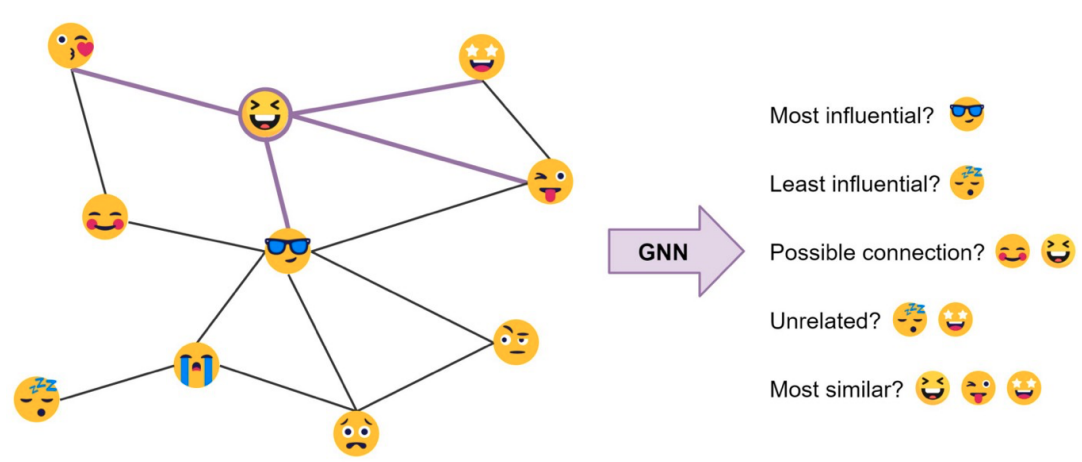
- Figure Credit: Chaitanya Joshi
Graph Classification






- Duvenaud et.al "Convolutional Networks on Graphs for Learning Molecular Fingerprints", NeurIPS 2015
How to construct GNN layers?
GNN Layers
- We construct permutation-equivariant functions $f(\mathbf{X}, \mathbf{A})$ over graphs by shared application of a local permutation-invariant $g(\mathbf{x}_i, \mathbf{X}_{\mathcal{N}_i})$.
- $f$: referred to as GNN layer
- $g$: referred to as "diffusion", "propagation", "message passing"
- How can we concretely define $g$:
- many designs have been proposed
- most proposed solutions fall into three spatial "flavors"
Three "Flavors" of GNN Layers



Convolutional GNN
- Features of neighbours aggregated with fixed weights $c_{ij}$ $$\mathbf{h}_i=\phi\left(\mathbf{x}_i, \bigoplus_{j\in\mathcal{N}_i}c_{ij}\psi(\mathbf{x}_j)\right)$$
- The weights depend directly on $\mathbf{A}$
- ChebyNet (Defferrard et al., NeurIPS 2016)
- GCN (Kipf & Welling, ICLR 2017)
- SGC (Wu et al., ICML 2019)
- Useful for homophilous graphs and scaling up:
- when edges encode label similarity

Attentional GNN
- Features of neighbours aggregated with implicit weights via attention $$\mathbf{h}_i=\phi\left(\mathbf{x}_i, \bigoplus_{j\in\mathcal{N}_i}a(\mathbf{x}_i,\mathbf{x}_j)\psi(\mathbf{x}_j)\right)$$
- Attention weights computed as $a_{ij}=a(\mathbf{x}_i,\mathbf{x}_j)$
- MoNet (Monti et al., CVPR 2017)
- GAT (Veličković, ICLR 2018)
- GaAN (Zhang et al., UAI 2018)
- Useful as "middle ground" w.r.t. capacity and scale:
- Edges need not encode homophily
- But still compute scalar value in each edge

Message-Passing GNN
- Compute arbitrary vectors ("messages") to be sent across edges $$\mathbf{h}_i=\phi\left(\mathbf{x}_i, \bigoplus_{j\in\mathcal{N}_i}\psi(\mathbf{x}_i, \mathbf{x}_j)\right)$$
- Messages computed as $\mathbf{m}_{ij}=\psi(\mathbf{x}_i, \mathbf{x}_j)$
- Interaction Networks (Battaglia et al., NeurIPS 2016)
- MPNN (Gilmer, ICML 2017)
- GraphNets (Battaglia et al., ICML 2018)
- Most generic GNN layer:
- May have scalability or learnability issues
- Ideal for computational chemistry, reasoning and simulation

Node Embeddings
Learning Node Embedding
- Early "successes" of deep learning on graphs was in embedding nodes into vectors $\mathbf{h}_u$ through an encoder.
- At the time, was implemeted as look-up table

What makes a node representation good?
- Graphs encode interesting structure.
- node representations must preserve graph structure
- Features of nodes $i$ and $j$ should be predictive of existence of edge $(i, j)$!
- Can be implemented through an unsupervised loss.
- $\mathbf{h}_i$ and $\mathbf{h}_j$ must be close iff $(i,j)\in\mathcal{E}$
- can use standard cross-entropy loss $$\sum_{(i,j)\in\mathcal{E}}\log \sigma\left(\mathbf{h}_i^T\mathbf{h}_j\right) + \sum_{(i,j)\notin\mathcal{E}}\log\left(1-\sigma(\mathbf{h}_i^T\mathbf{h}_j)\right)$$
Random-Walks on Graphs
- Link prediction objective is a special case of random-walk objectives
- redefine the condition from $(i,j)\in\mathcal{E}$ to $i$ and $j$ co-occur in a (short) random-walk
- Common unsupervised graph representation learning prior to GNNs
- DeepWalk (Perozzi et al., KDD'14)
- node2vec (Grover & Leskovec, KDD'16)
- LINE (Tang et al., WWW'15)
Local Objectives vs Conv-GNNs
- Random walk objectives inherently capture local similarities.
- A (convolutional) GNN also summarises local information of the graph!
- Neighbouring nodes tend to highly overlap in n-step neighbourhoods
- Conv-GNN enforces similar features for neighbouring nodes by design.
- DeepWalk-style models learn representations similar to that of convolutional GNN.
- Corollary 1: Random-walk objectives can fail to provide useful signal to GNNs.
- Corollary 2: At times, DeepWalk can be matched by an untrained Conv-GNN.
- Deep Graph Informax (ICLR 2019)
Transformers vs GNNs
Transformers
- Transformers is a special case of GCNs when the graph is fully connected.
- The neighborhood $\mathcal{N}_i$ is the whole graph.
$$\begin{eqnarray} h_i^{l+1} &=& W^lConcat_{k=1}^K(\sum_{j\in\mathcal{N}_i}\alpha_{ij}^{k,l}V^{k,l}h_j^l) \\ \alpha_{ij}^{k,l} &=& Softmax_{\mathcal{N}_i}(\hat{\alpha}_{ij}^{k,l}) \\ \hat{\alpha}_{ij}^{k,l} &=& Q^{l,k}h_i^lK^{l,k}h_j^l \end{eqnarray}$$

Transformers
- What does it mean to have a graph fully connected?
- It becomes less useful to talk about graphs as each data point is connected to all other points. There is no particular graph structure that can be used.
- It would be better to talk about sets rather than graphs in this case.
- Transformers are Set Neural Networks.
- They are today the best technique to analyze sets/bags of features.
Graph Transformers
- Graph version of Transformer: $$\begin{eqnarray} h_i^{l+1} &=& W^lConcat_{k=1}^K(\sum_{j\in\mathcal{N}_i}\alpha_{ij}^{k,l}V^{k,l}h_j^l) \\ \alpha_{ij}^{k,l} &=& Softmax_{\mathcal{N}_i}(\hat{\alpha}_{ij}^{k,l}) \\ \hat{\alpha}_{ij}^{k,l} &=& (Q^{l,k}h_i^l)^TK^{l,k}h_j^l \end{eqnarray}$$
- Attention mechanism in a one-hop neighborhood

Spectral GNNs
Convolution as Matrix Multiplication
- Consider convolution over a sequence represented as cyclical grid graph:
- Can we computed as a matrix multiplication with a circulant matrix s.t., $\mathbf{H}=\mathbf{CX}$ $$f(\mathbf{X})=\begin{bmatrix}b & c & & & a \\a & b & c & & \\ & \ddots & \ddots & \ddots & \\ & & a & b & c \\ c & & & a & b \end{bmatrix}\begin{bmatrix} - & \mathbf{x}_0 & - \\ - & \mathbf{x}_1 & - \\ - & \vdots & - \\ - & \mathbf{x}_{n-2} & - \\ - & \mathbf{x}_{n-1} & - \end{bmatrix}$$
Circulant Matrices: Properties and Eignevectors
- Circulant matrices commute
- $\mathbf{C}(\mathbf{v})\mathbf{C}(\mathbf{w})=\mathbf{C}(\mathbf{w})\mathbf{C}(\mathbf{v})$ for any vectors $\mathbf{v}$ and $\mathbf{w}$
- Matrices that commute are jointly diagonalisable.
- eigenvectors are shared across all such matrices
- Conveniently, the eigenvectors of circulants are the discrete Fourier basis: $$\phi_{l} = \frac{1}{\sqrt{n}}\left(1,e^{\frac{2\pi il}{n}},e^{\frac{4\pi il}{n}},\dots,e^{\frac{2\pi i(n-1)l}{n}}\right)^T \quad l=0,1,2,\dots,n-1$$
- This can be easily computed by studying $\mathbf{C}([0, 1, 0, 0, 0, \dots])$, which is the shift matrix.
DFT and Convolution
- We can eigendecompose any circulant matrix as $\mathbf{C}(\Lambda)=\Phi\Lambda\Phi^*$
- $\Phi$ are the Fourier basis
- diagonal values of $\Lambda$ are eignevalues
- Convolution theorem naturally follows: $$f(\mathbf{X})=\mathbf{C}(\Lambda)\mathbf{X}=\Phi\Lambda\Phi^*\mathbf{X}=\Phi\begin{bmatrix}\hat{\lambda}_0 & & \\ & \ddots & \\ & & \hat{\lambda}_{n-1} \end{bmatrix}\Phi^*\mathbf{X}=\Phi(\hat{\lambda} \odot \hat{X})$$
- If $\Phi$ is known, we can express convolution in terms of $\hat{\lambda}_i$ rather than spatial parameters $\mathbf{\theta}$.
Convolutions on Graphs
- On graphs, convolutions of interest need to be more generic than circulants.
- We can still use the concept of joint/common eigenbases.
- If we know a "Fourier basis for graphs", $\Phi$, only focus on learning eigenvalues.
- For grids, we wanted our convolutions to commute with shifts.
- With adjacency matrix generalize to non-grids (shift matrix is special case for grid graph).
- For convolution on regular grid of $n$ nodes, $\Phi$ was always the same (n-way DFT).
- Now every graph will have its own $\Phi$.
- Want our convolution to commute with A, but we cannot always eigendecompose A.
- Instead, use the graph Laplacian matrix, $\mathbf{L} = \mathbf{D} - \mathbf{A}$, where $\mathbf{D}$ is the degree matrix.
- Captures all adjacency properties in a mathematically convenient way.
Graph Laplacian

$$\mathbf{L}=\begin{bmatrix}2 & -1 & 0 & 0 & -1 & 0 \\-1 & 3 & -1 & 0 & -1 & 0 \\0 & -1 & 2 & -1 & 0 & 0 \\ 0 & 0 & -1 & 3 & -1 & -1 \\ -1 & -1 & 0 & -1 & 3 & 0 \\ 0 & 0 & 0 & -1 & 0 & 1\end{bmatrix}$$
Graph Fourier Transform
- For undirected graphs, $\mathbf{L}$ is:
- Symmetric
- Positive semi-definite
- easy to eigendecompose
- We can express $\mathbf{L}=\Phi\Lambda\Phi^*$
- Changing the eigenvalues in $\Lambda$ expresses any operation that commutes with $\mathbf{L}$.
- Commonly referred to as the "graph Fourier transform" (Bruna et al., ICLR'14)
- To convolve with a feature matrix $\mathbf{X}$, we do the following (learn $\lambda_i$): $$f(\mathbf{X})=\Phi\begin{bmatrix}\hat{\lambda}_0 & & \\ & \ddots & \\ & & \hat{\lambda}_{n-1} \end{bmatrix}\Phi^*\mathbf{X}$$
Spectral GNNs
- Directly learning eigenvalues is typically inappropriate:
- Not localized, doesn't generalize to other graphs, computationally expensive, etc.
- Common solution: make eigenvalues $\mathbf{\Lambda}$ related to eigenvalues of $\mathbf{L}$
- Typically by a degree-$k$ polynomial function, $p_k$
- Yielding $f(x)=\mathbf{\Phi}p_k(L)\mathbf{\Phi^*x}$
- Common variants:
- Cubic splines (Bruna et al., ICLR'14)
- Chebyshev polynomials (Defferrard et al., NeurIPS'16)
- Cayley polynomials (Levie et al., Trans. Sig. Proc.'18)
- Using a polynomial in $\mathbf{L}$ we can define a conv-GNN
- With coefficients defined by $c_{ij}= (p_k(L))_{ij}$
Transformers with Positional Encoding for Graphs
- Transformers are typically defined on sets.
- Order information can be provided to transformers through positional embeddings
- Sines/cosines sampled depending on position $$\begin{equation} PE_{pos, 2i} = sin\left(\frac{pos}{10000^{\frac{2i}{d_{emb}}}}\right) \mbox{ and } PE_{pos, 2i+1} = cos\left(\frac{pos}{10000^{\frac{2i}{d_{emb}}}}\right) \end{equation}$$
- Very similar to the DFT eigenvectors.
- Positional embeddings can be interpreted as eigenvectors of the grid graph.
- Same idea can be leveraged to define Transformers on Graphs
- Just feed some eigenvectors of the graph Laplacian (columns of $\Phi$)
- Graph Transformer from Dwivedi & Bresson (2021)
Probabilistic Graphical Models
Probabilistic Modeling
- Treat graphs as probabilistic graphical models (PGMs)
- Nodes: Random variables
- Edges: dependencies between distributions
- Encode independence and conditional independence
- allows for compact representation of multivariate probabilistic distributions
 $$p(a,b,c,d,e) = p(a)p(b)p(c|a,b)p(d|e)p(c|e)$$
$$p(a,b,c,d,e) = p(a)p(b)p(c|a,b)p(d|e)p(c|e)$$
Markov Random Fields
- A particular PGM of interest is Markov Random Feild (MRF)
- allows decompositio of joint distribution into product of edge potentials
- MRFs are modelled through inputs $\mathbf{X}$ and latents $\mathbf{H}$.
- latents are akin to node representations
- inputs and latents for each node are are related to each other independently
- latents are related accordingly to the edges of the graph
- Joint probability distribution: $$p\left(\mathbf{X},\mathbf{H}\right)=\prod_{v\in\mathcal{V}}\phi\left(\mathbf{x}_v,\mathbf{h}_v\right)\prod_{(v_1,v_2)\in\mathcal{E}}\psi\left(\mathbf{h}_{v_1},\mathbf{h}_{v_2}\right)$$
- $\phi(\cdot,\cdot)$ and $\psi(\cdot,\cdot)$ are real-valued potential functions
Mean-Field Inference
- To embed nodes, we need to sample from the posterior, $p(\mathbf{H}|\mathbf{X})$
- estimating the posterior is intractable, even if exact potential functions are known
- One popular method of resolving this is mean-field variational inference
- Assume posterior is approximated by a product of node-level densities $$p(\mathbf{H}|\mathbf{X})\approx \prod_{v \in \mathcal{V}}q(\mathbf{h}_v)$$
- $q(\cdot)$ is a density that is easy to compute and sample from, e.g., Gaussian
- $q(\cdot)$ can be estimated by minimizing $KL\left(\prod_{v}q(v|X)\|p(\mathbf{H}|\mathbf{X})\right)$ to true posterior
- Minimising the KL is intractable, but it admits a favourable approximate algorithm
Mean-Field Inference as GNN
- We can iteratively update $q(\cdot)$ starting from an initial guess $q^{(0)}(\mathbf{h})$ (variational inference): $$\log q^{(t+1)}(\mathbf{h}_i) = c_i + \log\phi(\mathbf{x}_i,\mathbf{h}_i) + \sum_{j\in\mathcal{N}_i}\int_{\mathbb{R}^k}q^t(\mathbf{h}_i)\log\psi(\mathbf{h}_i,\mathbf{h}_j)d\mathbf{h}_i$$
- Aligns nicely with message-passing GNN. $$\mathbf{h}_i = \phi\left(\mathbf{x}_i,\bigoplus_{j\in\mathcal{N}_i}\psi(\mathbf{x}_i, \mathbf{x}_j)\right)$$
Graph Isomorphism Testing
How powerful are Graph Neural Networks?
- GNNs are a powerful tool for processing real-world graph data
- But they won't solve any task specified on a graph accurately!
- Canonical example: deciding graph isomorphism
- Can a GNN distinguish between two non-isomorphic graphs i.e., $\mathbf{h}_{G_1} \neq \mathbf{h}_{G_2}$?
- Permutation invariance mandates that two isomorphic graphs will always be indistinguishable, so this case is OK.
Weisfeiler-Leman Test
- Simple but powerful way of distinguishing: pass random hashes of sums along the edges
- Connection to conv-GNNs spotted very early; e.g., by GCN (Kipf & Welling, ICLR'17)
- It explains why untrained GNNs work well!
- Untrained $\sim$ random hash
- The test can fail sometimes

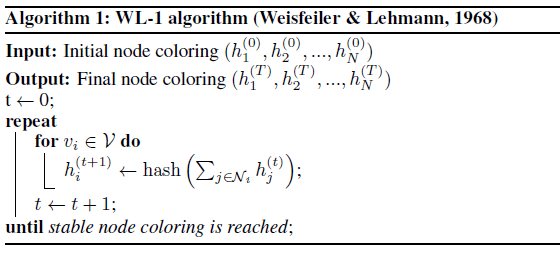
GNNs are no more powerful than 1-WL
- Over discrete features, GNNs can only be as powerful as the 1-WL test described before
- One important condition for maximal power is an injective aggregator (e.g. sum)
- Graph isomorphism network (GIN; Xu et al., ICLR'19) proposes a simple, maximally-expressive GNN, following this principle $$h_v^{(k)}=MLP^{(k)}\left(\left(1+\epsilon^{(k)}\right)\cdot h_v^{(k-1)}+\sum_{v \in \mathcal{N(v)}}h_v^{(k-1)}\right)$$
Higher-Order Graph Neural Networks
- We can make GNNs stronger by analysing failure cases of 1-WL.
- For example, just like 1-WL, GNNs cannot detect closed triangles
- Augment nodes with randomised/positional features (Murphy et al., ICML'19 and You et al., ICML'19)
- Can also literally count interesting subgraphs (Bouritsas et al., 2020)
- k-WL labels subgraphs of k nodes together.
- Exploited by 1-2-3-GNNs (Morris et al., AAAI'19)
- If features are real-valued, injectivity of sum falls apart.
- No aggregator is better than another aggregator.
Geometric Deep Learning
Geometric Deep Learning
- GNNs were described in terms of invariances and equivariances.
- Many standard architectures like CNNs and Transformers can be recoverd by,
- Local and equivariant layer specified over neighbourhoods
- Activation functions
- (Potentially: pooling layers that coarsen the structure)
- Global and invariant layer over the entire domain
- Can design a more general class of geometric deep learning architectures.
"Five Gs" of Geometric Deep Learning
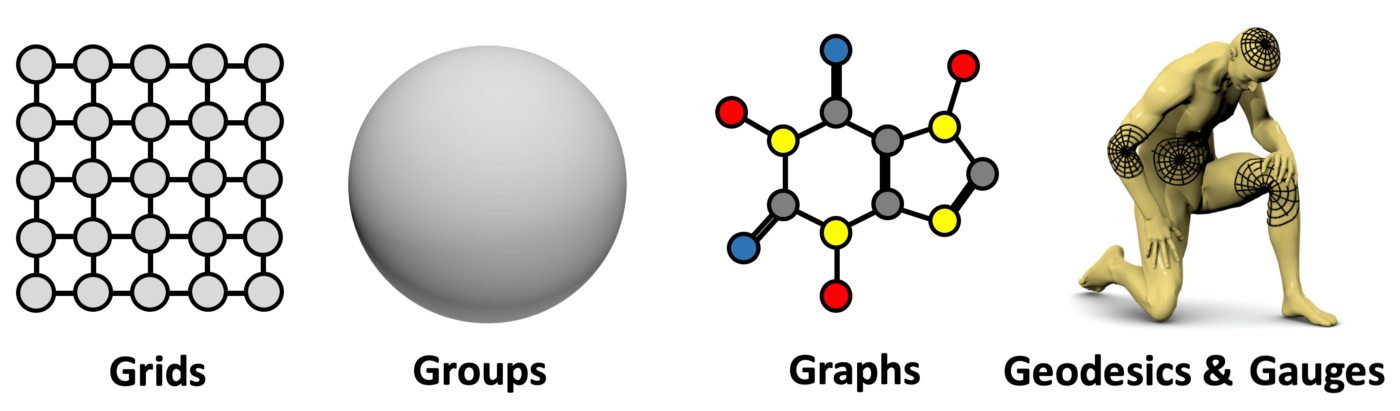
Applications
Quantum Chemistry
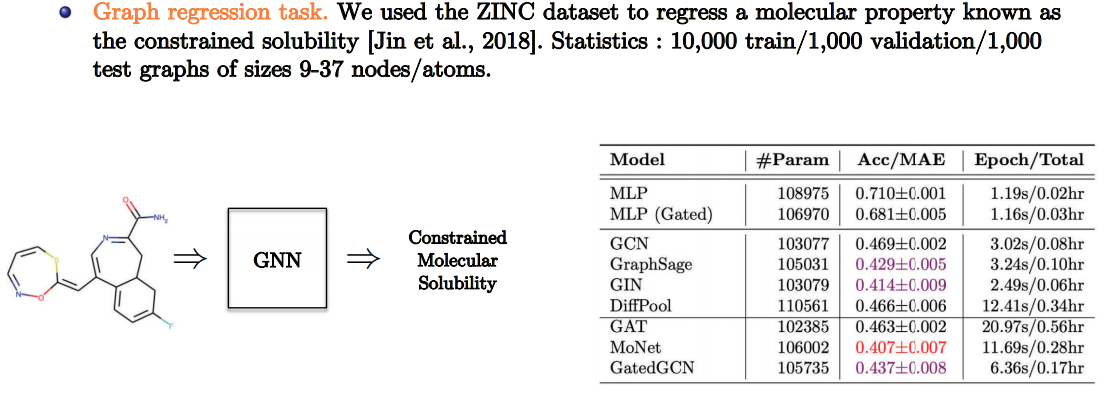
- Slide Credit: Xavier Bresson
Computer Vision
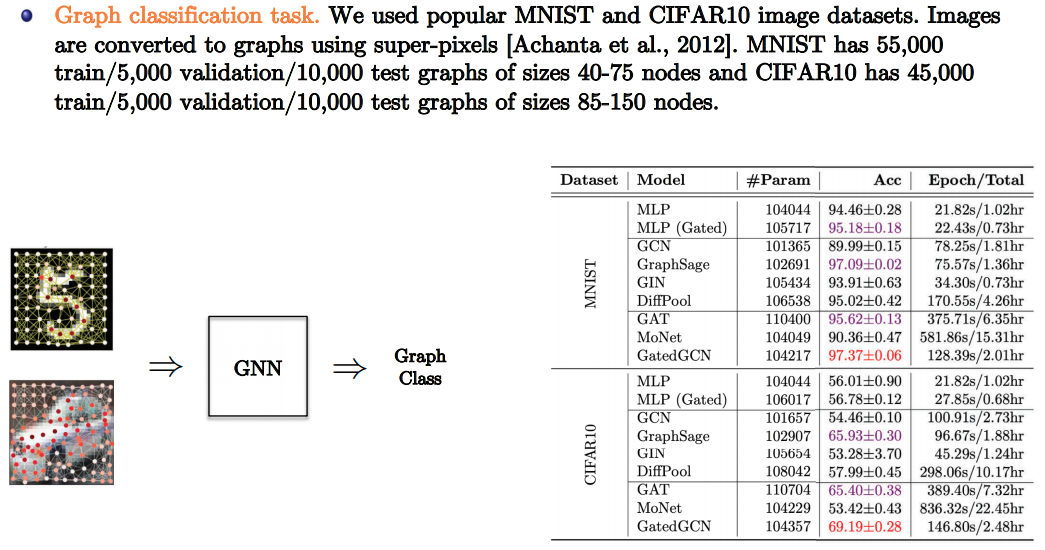
- Slide Credit: Xavier Bresson
Combinatorial Optimization
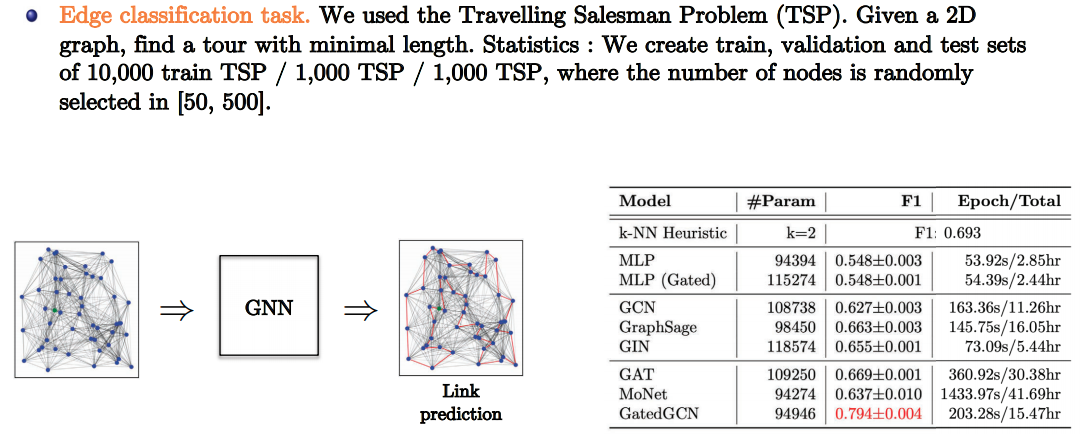
- Slide Credit: Xavier Bresson
Semi-Supervised Classification on Graphs
- Setting:
- Some nodes are labeled
- All other nodes are unlabeled
- Task:
- Predict node label of unlabeled nodes
- Evaluate loss on labeled nodes only: $$ \begin{equation} \mathcal{L} = -\sum_{l\in\mathcal{Y}_L}\sum_{f=1}^F Y_lf \log Z_lf \end{equation} $$

- $\mathcal{Y}_L$: set of labled node indices
- $\mathbf{Y}$: label matrix
- $\mathbf{Z}$: GCN output (after softmax)
Normalization and Depth
- What kind of normalization can we do for graphs?
- Node normalization
- Pair normalization
- Edge normalization
- Does depth matter for GCNs?

Scalable Inception Graph Neural Networks
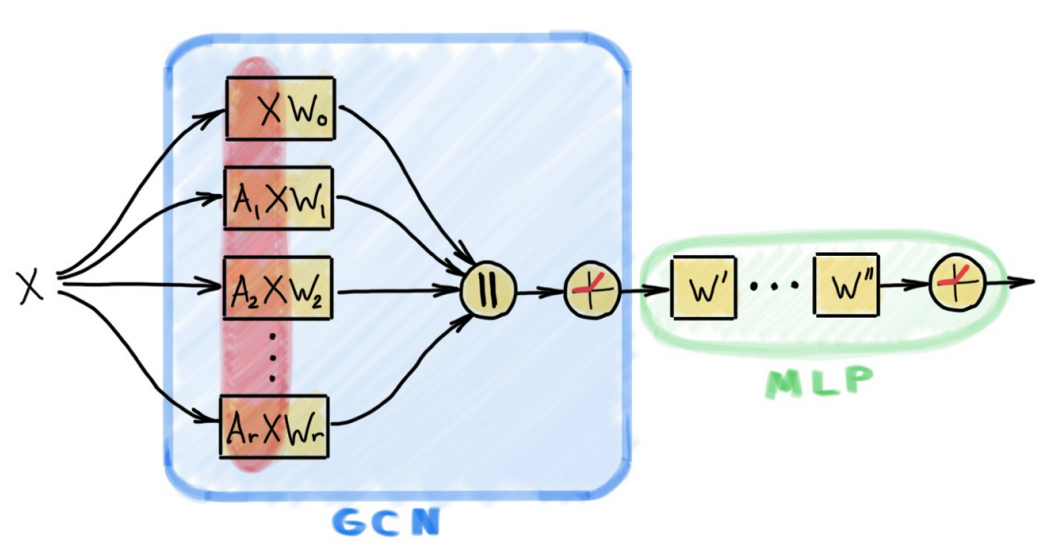
- Figure Credit: Michael Bronstein
Scalable Inception Graph Neural Networks


Graph CNN Challenges
- How to define compositionality on graphs (i.e. convolution, downsampling, and pooling on graphs)?
- How to ensure generalizability across graphs?
- And how to make them numerically fast? (as standard CNNs)
- Scalability: millions of node, billions of edges
- How to process dynamic graphs?
- How to incorporate higher-order structures?
- How powerful are graph neural networks?
- limited theoretical understanding
- message passing based GCN are no stronger than Weisfeiler-Lehman tests.
Getting Started
- Lots of open questions and uncharted territory.
- Standardized Benchmarks: Open Graph Benchmark
- Software Packages: Pytorch Geometric or Deep Graph Library
