Abstract
Matching of deformed patterns is an important and difficult task in biometric recognition. Moreover, recognition becomes considerably more difficult in 1 : 1 matching schemes where only a single probe and a single gallery sample are available to determine a match. Referred to as Probabilistic Deformation Models (PDMs), an approach originally proposed by Thornton et al. for matching iris images reduces the image matching problem to matching local image regions, where iris distortions are approximated by local independent spatial translations and then related by a Gaussian Markov Random Field (GMRF). Building on Thornton’s method, this paper demonstrates the drawbacks of using too simple of a model to capture biometric deformations when restricted to 1 : 1 matching schemes. In this paper we propose several improvements, both from a computational as well as a performance point of view, to the basic framework. The new model, which extends PDM (referred to as ‘ePDM’), allows us to capture a more varied set of valid pattern deformations for authentic matches that are present in biometric signatures captured in 1 : 1 matching environments. We demonstrate the effectiveness of this model via extensive numerical results on multiple biometric databases while comparing to other state-of-the-art 1 : 1 matching algorithms.
Overview
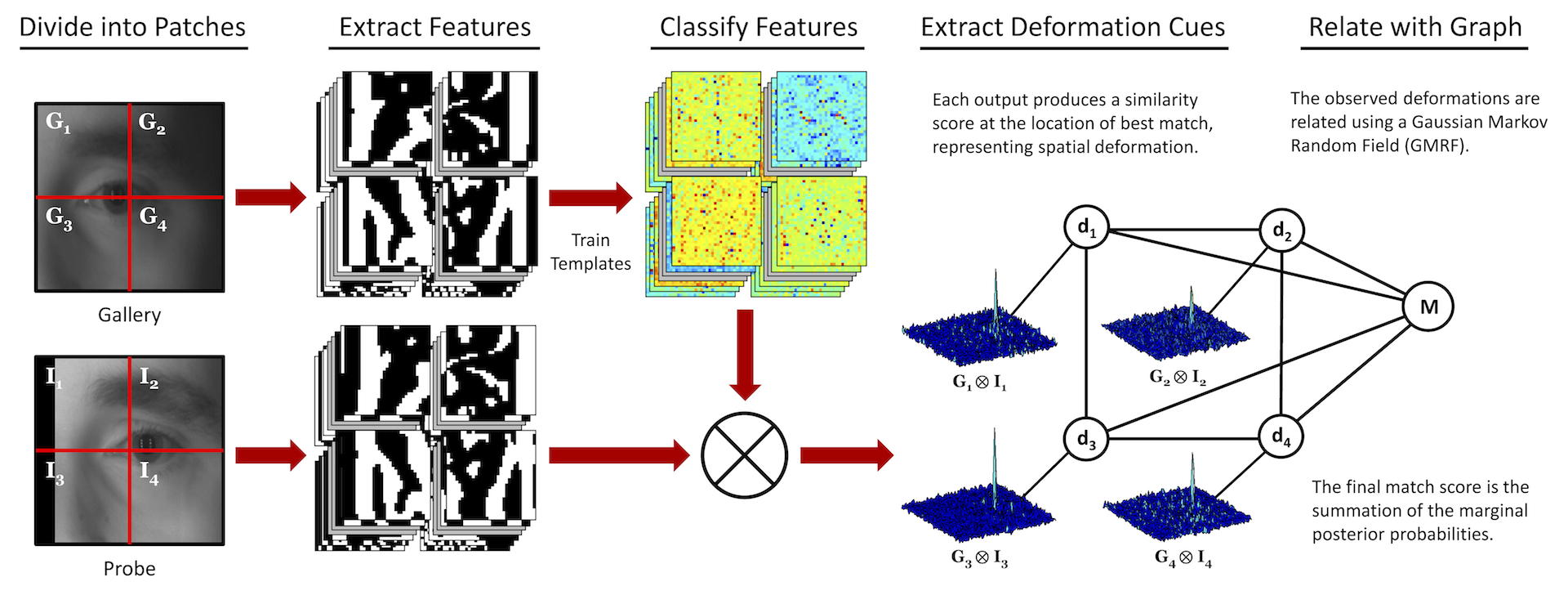
System overview for determining a match. The probe, \(\textbf{I}\), and gallery, \(\textbf{G}\), images are each divided into non-overlapping patches where the corresponding probe and gallery patches are compared via template matching. The outputs from template matching are then used as inputs into the GMRF model that is trained to capture the relationship between the deformations of the image patches for authentic matches. The final match score \(M\) is the summation of the marginal posterior probabilities.
Contributions
- Introduce several algorithmic improvements to the original PDM model to improve deformation tolerance in different biometric modalities and 1 : 1 matching schemes.
- Improve the memory and computational complexity of both the training and testing procedures proposing a computationally efficient classifier design.
- A comprehensive comparison to several other methods on periocular datasets demonstrating the need for deformation estimation in 1 : 1 matching schemes as well as the efficacy of PDM. We achieve state-of-the-art results on multiple databases for periocular verification of varying difficulty.
References
-
Jonathon M. Smereka, Vishnu Naresh Boddeti, B.V.K. Vijaya Kumar and Andres Rodriguez, Stacked Correlation Filters for Biometric Verification, IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP 2016)
-
Jonathon M. Smereka, Vishnu Naresh Boddeti and B.V.K. Vijaya Kumar, Probabilistic Deformation Models for Challenging Periocular Image Verification, IEEE Transactions on Information Forensics and Security, 2015
-
Jonathon M. Smereka and B.V.K. Vijaya Kumar, What is a ‘Good’ Periocular Region for Recognition? IEEE CVPR Workshop on Biometrics, June 2013
-
Arun Ross, Raghavender Jillela, Jonathon M. Smereka, Vishnu Naresh Boddeti, B. V. K. Vijaya Kumar, Ryan Barnard, Xiaofei Hu, Paul Pauca, Robert Plemmons, Matching Highly Non-Ideal Ocular Images: An Information Fusion Approach, 5th IAPR International Conference on Biometrics, 2012 (Oral)
-
Vishnu Naresh Boddeti, B.V.K. Vijaya Kumar and Krishnan Ramkumar, Improved Iris Segmentation Based on Local Texture Statistics, 45th Asilomar Conference on Signals, Systems and Computers, 2011 (Oral, Invited Paper)
-
Vishnu Naresh Boddeti, Jonathon M. Smereka and B.V.K. Vijaya Kumar, A comparative evaluation of iris and ocular recognition methods on challenging ocular images International Joint Conference on Biometrics, 2011. (Oral)
-
Jason Thornton, Marios Savvides and B.V.K. Vijaya Kumar, A Bayesian Approach to Deformed Pattern Matching of Iris Images, IEEE Transactions on Pattern Analysis and Maching Intelligence, 2007